There are many ways to look at the relationship between two categorical variables.
A simple contingency table will often do the job.
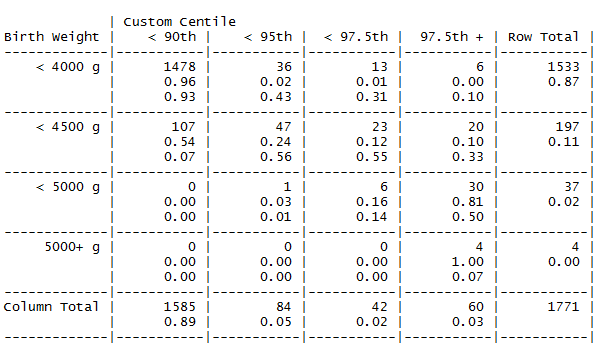
library(gmodels)
macroTable <- (data$macro, data$macroCust,
dnn = c("Birth Weight", "Customised Centile")
CrossTable(macroTable, digits = 2, prop.t = F, prop.chisq = F)
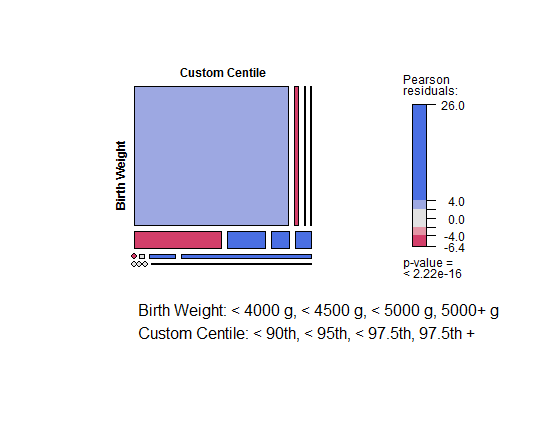
Mosaic plots are pretty cool.
library(vcd)
mosaic(macroTable,
labeling = labeling_list(sep = ", ", cols = 1),
margins = c(bottom = 7),
legend = T,
shade = T)
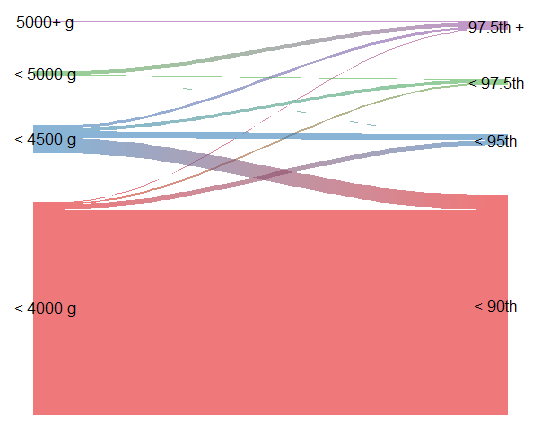
However, something I’ve really come to appreciate is the river plot. For example:
You can make these using the R library(riverplot), but you have to do a bit of data wrangling first. I have written a function to help with this when I am working from a data frame where the two factor variables are in separate columns, and the rows are observations.
data <- data.frame("ID" = c(1:500),
"A" = factor(sample(c(1:4), 500, replace = T),
labels = c("A", "B", "C", "D")),
"B" = factor(sample(c(1:3), 500, replace = T),
labels = c("Big", "Mid", "Small")))
makeRivPlot(data, "A", "B")
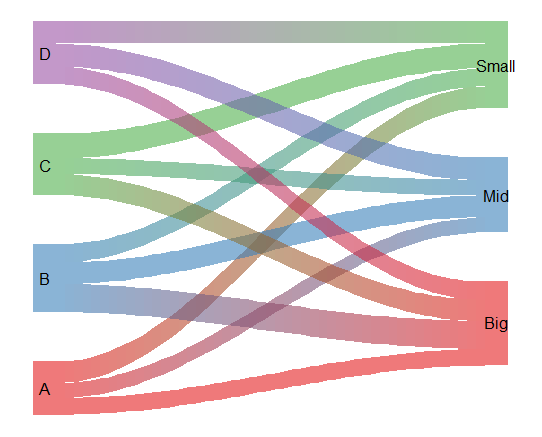
Here is the function.
makeRivPlot <- function(data, var1, var2) {
require(dplyr) # Needed for the count function
require(riverplot) # Does all the real work
require(RColorBrewer) # To assign nice colours
names1 <- levels(data[, var1])
names2 <- levels(data[, var2])
var1 <- as.numeric(data[, var1])
var2 <- as.numeric(data[, var2])
edges <- data.frame(var1, var2 + max(var1, na.rm = T))
edges <- count(edges)
colnames(edges) <- c("N1", "N2", "Value")
nodes <- data.frame(
ID = c(1:(max(var1, na.rm = T) +
max(var2, na.rm = T))),
x = c(rep(1, times = max(var1, na.rm = T)),
rep(2, times = max(var2, na.rm = T))),
labels = c(names1, names2) ,
col = c(brewer.pal(max(var1, na.rm = T), "Set1"),
brewer.pal(max(var2, na.rm = T), "Set1")),
stringsAsFactors = FALSE)
nodes$col <- paste(nodes$col, 95, sep = "")
river <- makeRiver(nodes, edges)
return(plot(river))
}
Related Links: